What WE do
Next Generation Sequencing
Services
Explore the Sequencing Revolution with SeqPal – Your Genomic Maestro! We offer expert Next-Gen Sequencing (NGS) services, from library prep to cutting-edge sequencing and in-depth bioinformatics.
Tailor your genomics journey with SeqPal, letting us handle complexities while you focus on groundbreaking solutions. Elevate your genomics game with expertise, innovation, and personalized precision – choose SeqPal!
WHAT SEQPAL CAN DO
NGS Services
Targeted Sequencing
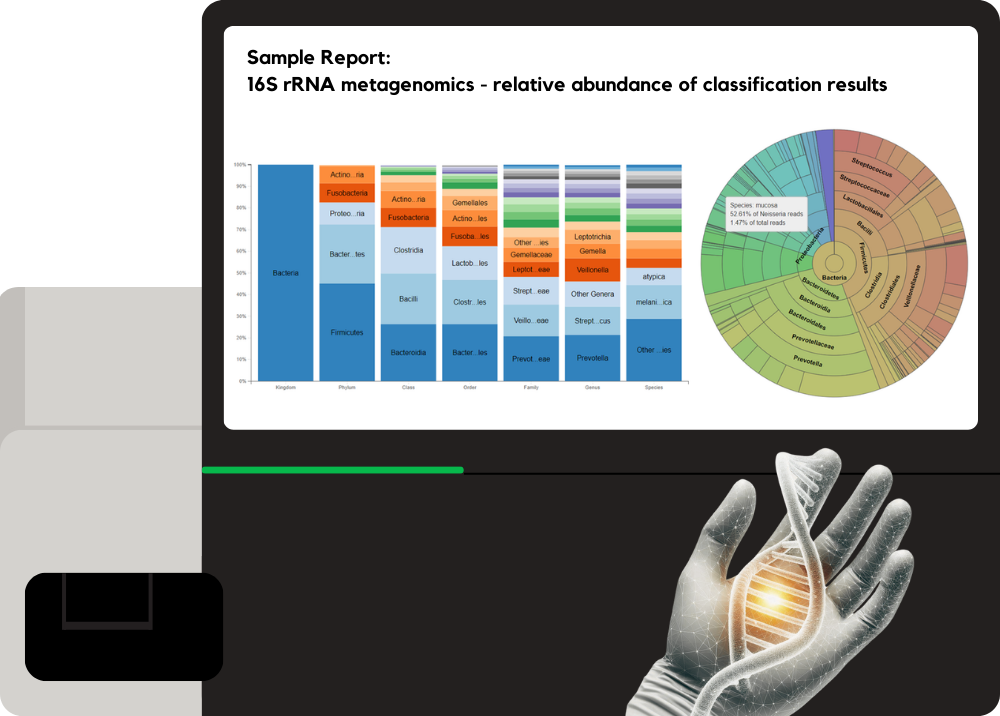
Metagenomics 16S ITS
CRISPR validation
Whole Genome Sequencing (WGS)
Whole Exome Sequencing (WES)
RNA Sequencing
CHIP Sequencing
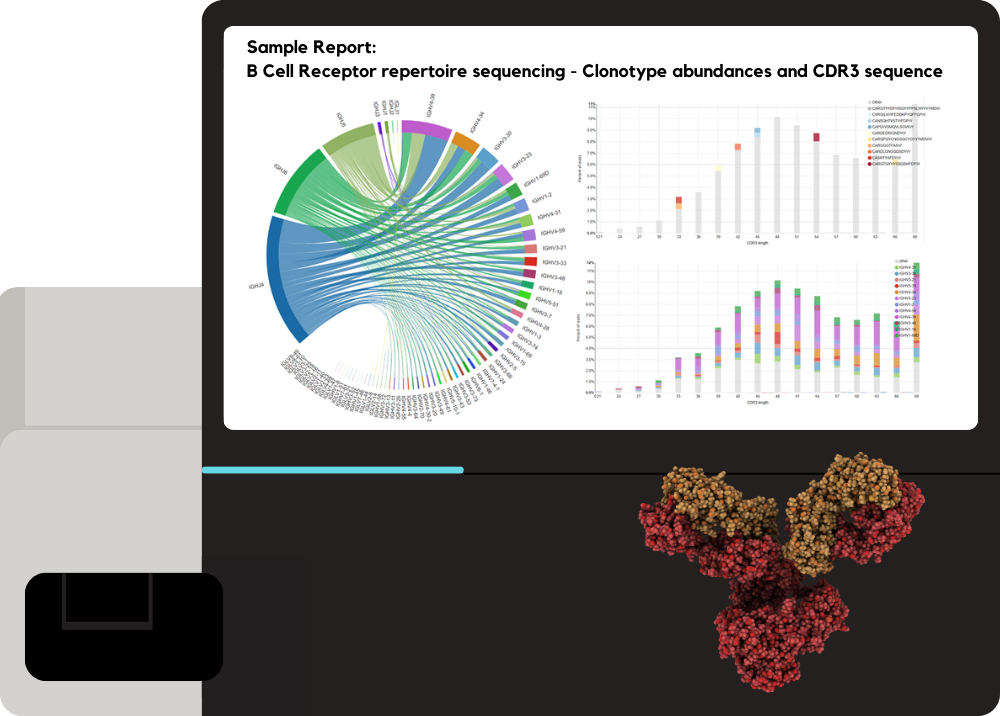
Single-cell Sequencing
Long-read Nanopore Sequencing
Methyl sequencing
ATAC Sequencing
Complete GENOMICS
Why Seqpal
SeqPal Advantage
Tailored workflows to meet your specific needs

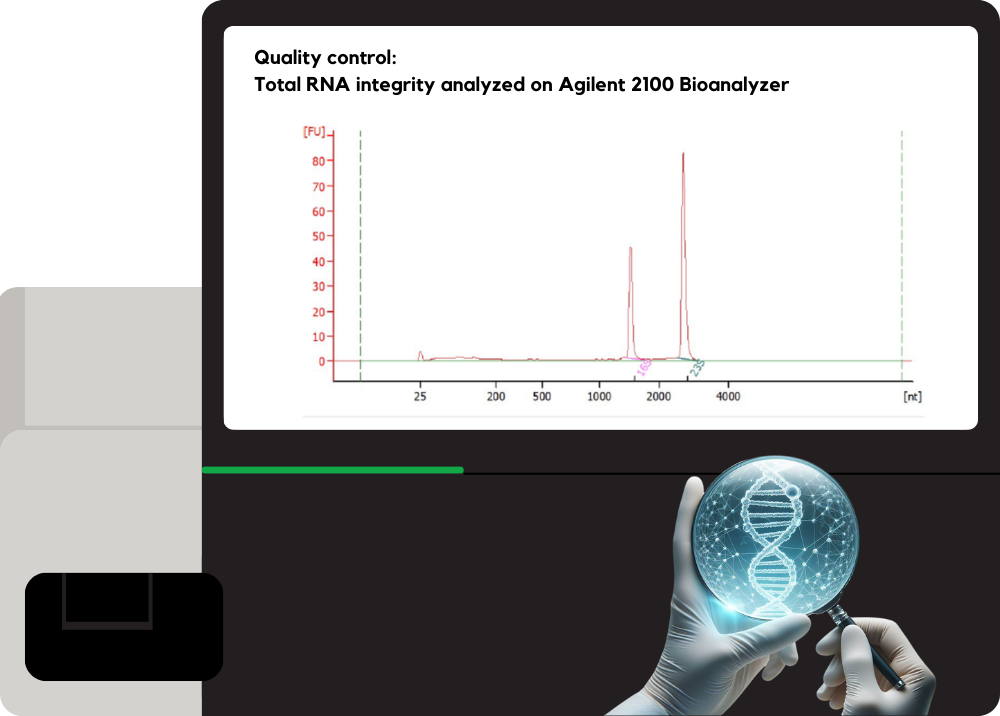
Quality Library Prep
SeqPal provides quality-controlled library preparation services utilizing kits from leading brands.
Affordable Services
Affordable, high-quality NGS services partner in San Francisco Bay Area.
Quick Turnaround Time
Submit your libraries and receive a bioinformatics report in as little as 3-5 days.
FAQs
What if my sample concentration is lower than the recommended concentration?
Different library prep kits and sequencer systems have varied minimum requirements. Please contact us, info@seqpal.com, to discuss the minimum amount of material needed for your specific kit or system.
Are you able to perform DNA or RNA extraction?
Certainly! We offer DNA and RNA extraction services. Reach out to us to discuss your sample type and requirements.
Is there any other information that needs to be sent before sending the samples?
DNA can be shipped in ambient conditions, while RNA requires shipment in dry ice for optimal quality. Please adhere to these guidelines for sample transportation.
What kit or library preparation protocol is used?
We primarily use Illumina’s kits for DNA-seq and mRNA-seq. However, we can accommodate other vendors’ kits upon request. Feel free to contact us for more information on library preparation.
What primers are used for 16S sequencing?
We use primers for the V3 and V4 hypervariable regions of 16S rRNA. If you have specific primer or region requirements, contact us to discuss custom protocols.
How many reads do I need for my samples?
The number of reads required varies based on your sample type and research goals. Here are general guidelines:
- Whole Genome Sequencing: Aim for 30X coverage.
- RNA-seq:
- Small Genomes: 5-10 million reads per sample.
- Human or Mouse: Target 20-30 million reads per sample.
- De novo transcriptome assembly projects may require 100 million reads per sample.
- Whole Exome Sequencing: Optimal coverage is in the range of 100X-200X.
- Bisulfite Sequencing: Aim for 30X coverage.
These guidelines serve as a starting point, and we recommend reaching out to us for a more personalized assessment based on your specific sample characteristics and research objectives.
How do I calculate coverage?
Coverage is calculated using the formula: Coverage = (read length) x (total number of reads) / (genome size). For example, 30x coverage of the human genome requires 600 million reads of 150 bp.
What if my sequencing needs do not fill up a lane or a flow cell?
If your sequencing requirements do not utilize the full capacity of a lane or flow cell, we offer the option to pool your libraries or samples with compatible ones. Please get in touch with us to discuss and arrange the appropriate pooling strategy that suits your needs.
Quote